nnU-Net
Automatic configuration and training of U-Net-based segmentation pipelines. Works out-of-the-box for a broad range of datasets from all imaging domain! Supports 2D and 3D (multi-channel) images.
Cite this software
Description
What is nnU-Net about?
When confronted with a new dataset, the application of AI-based segmentation methods is non-trivial. Substantial manual effort is required to optimize the network topology, preprocessing, training procedure and inference strategy to the task at hand. Such adaptations are non-trivial and require substantial expertise to ensure optimal performance on each dataset.
Consequently, access to state-of-the-art segmentation methods is often barred to domain researchers who are looking to analyze their data.
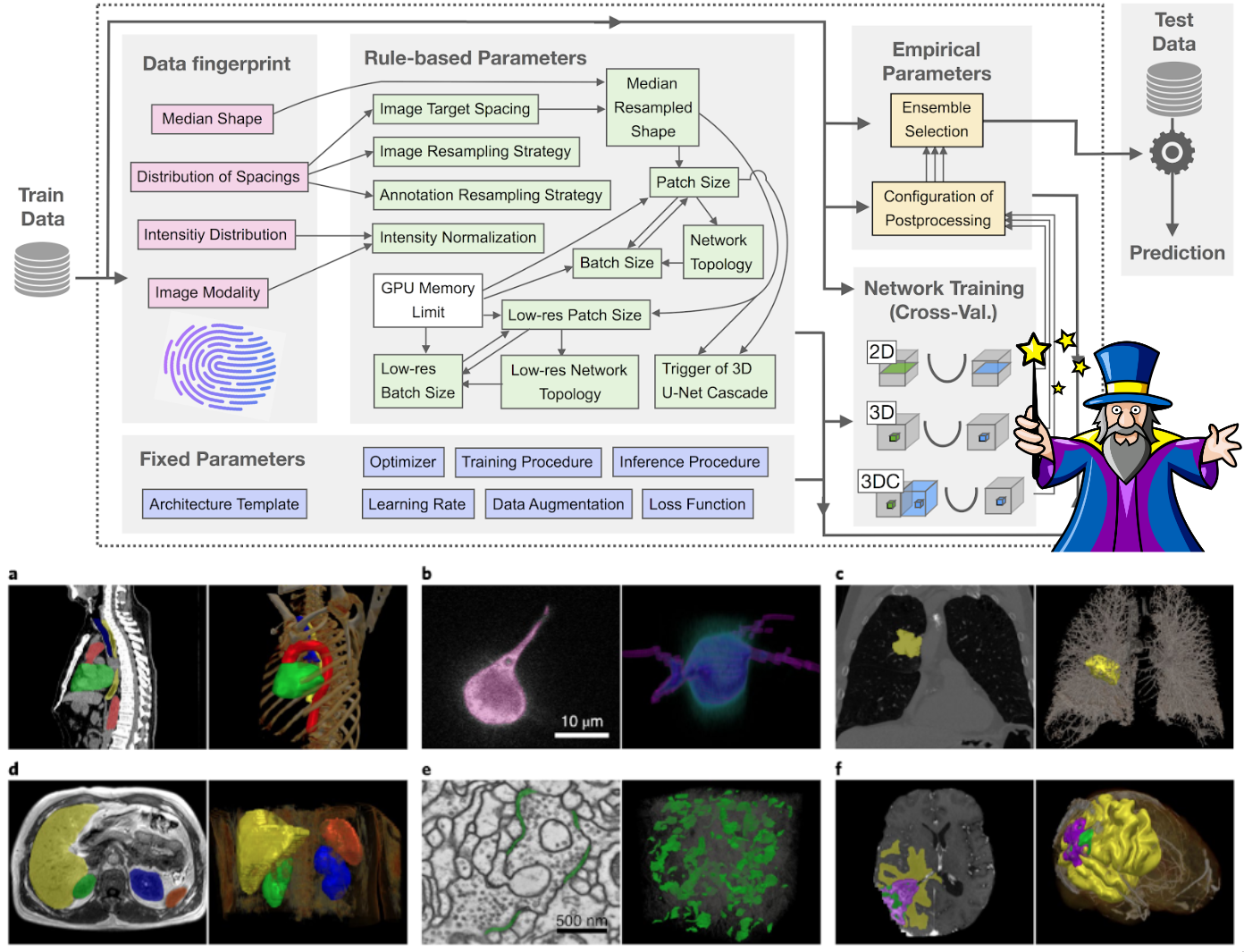
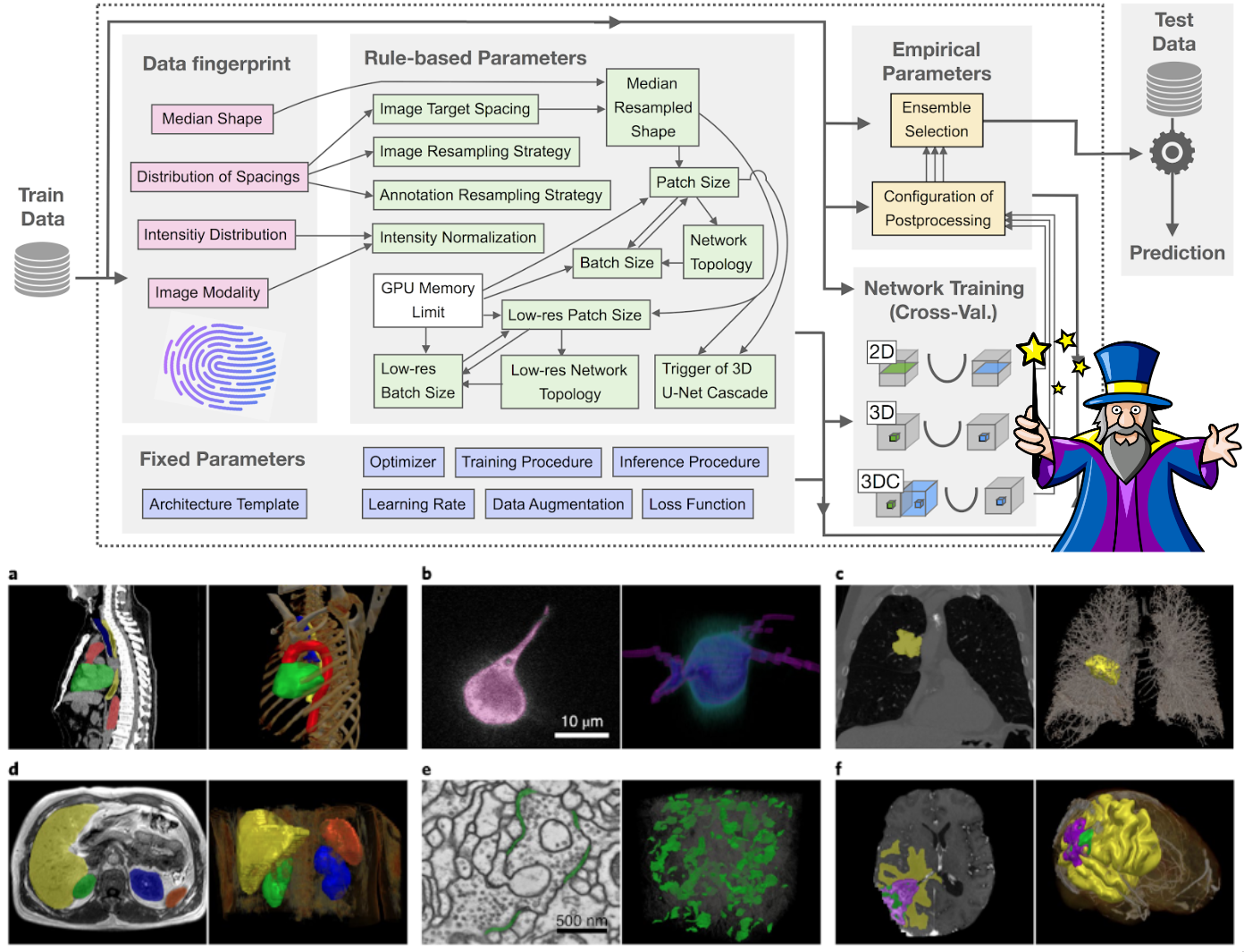
nnU-Net systematizes and automates this process, enabling it to be applied out-of-the-box to a wide range of datasets from many different imaging domains. All that is needed to apply nnU-Net is an annotated training dataset. nnU-Net does the rest: at the end of its automated pipeline it will return fully configured and trained U-Net models that can be used to run predictions on new images.
nnU-Net is able to work with both 2D and 3D data. It particularly shines in 3D: we applied it to 23 segmentation competitions with open leaderboards where it placed first in 33/55 segmentation tasks!
nnU-Net's core principle revolves around this differentiation of method parameters into three categories: fixed parameters that do not require adaptations between datasets, parameters that can be inferred from the properties of the presented data (such as image shapes and voxel spacings) as well as empirical parameters. Have a look at our Nature Methods paper to learn more!
Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J., & Maier-Hein, K. H. (2021). nnU-Net: a self-configuring
method for deep learning-based biomedical image segmentation. Nature methods, 18(2), 203-211.
Access and Usage Instructions
You can find nnU-Net along with installation & usage instructions here: https://github.com/MIC-DKFZ/nnUNet
Acknowldegements

nnU-Net is developed and maintained by the Applied Computer Vision Lab (ACVL) of Helmholtz Imaging and the Division of Medical Image Computing at the German Cancer Research Center (DKFZ).
- Apache-2.0
Participating organisations
Contributors
Contact person
Fabian Isensee
Lead Developer & Inventor
Helmholtz Imaging, German Cancer Research Center (DKFZ)
0000-0002-3519-5886
Mail FabianRelated projects
Helmholtz Imaging
We catalyze scientific discovery from sensory measurements to knowledge.