CellDetection
An open-source Python package, offering advanced AI methods for the identification and segmentation of biomedical objects, such as cells, in image data. Includes trained models that work out-of-the-box for a wide range of use cases.
Description
Cell Detection
⭐ Showcase
NeurIPS 22 Cell Segmentation Competition
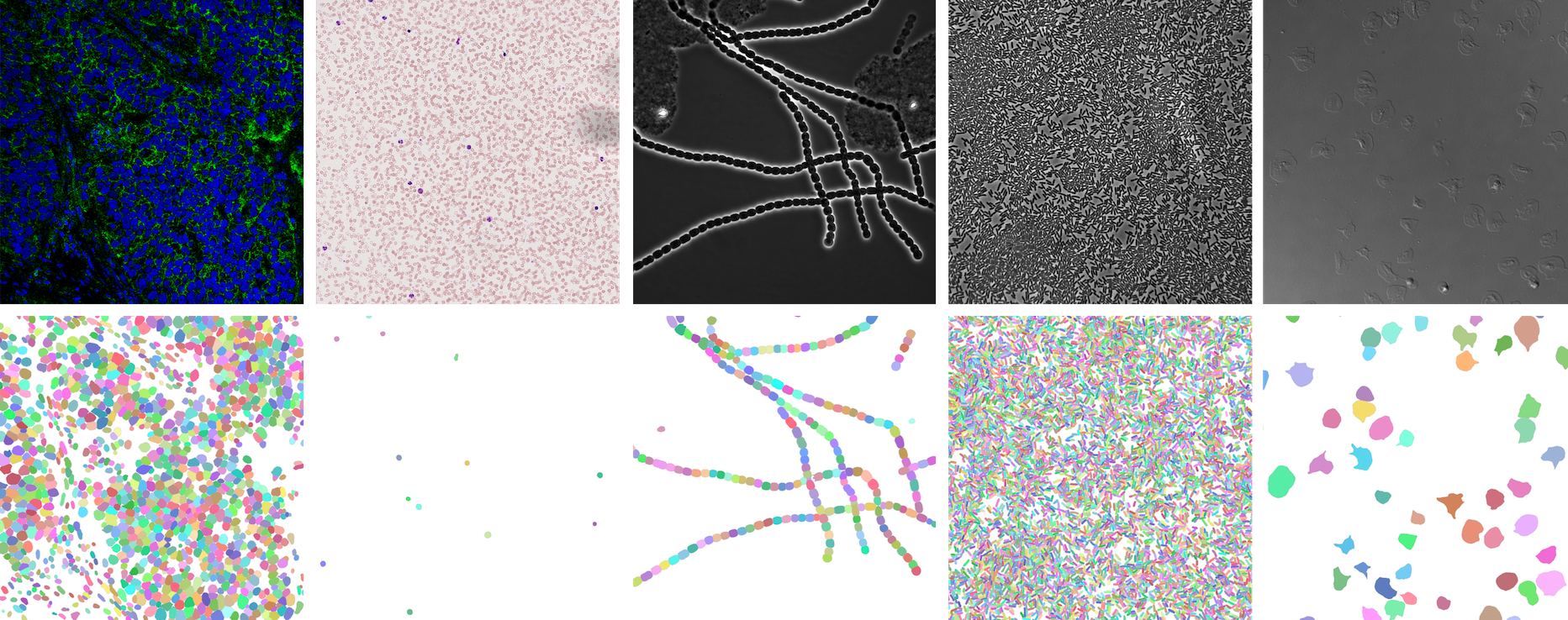
https://openreview.net/forum?id=YtgRjBw-7GJ
Nuclei of U2OS cells in a chemical screen
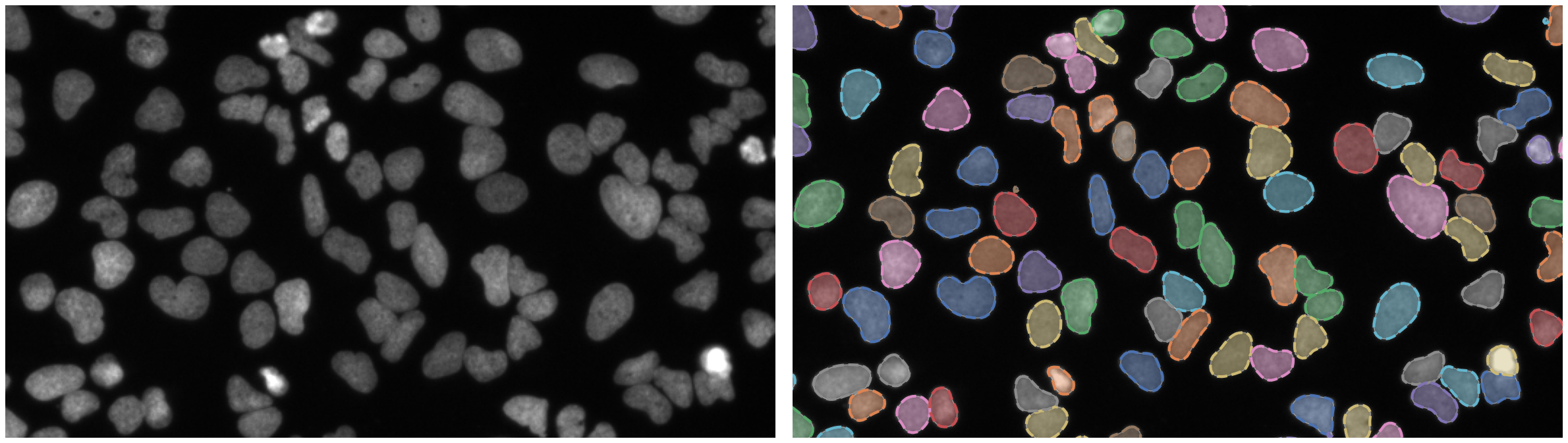
https://bbbc.broadinstitute.org/BBBC039 (CC0)
P. vivax (malaria) infected human blood
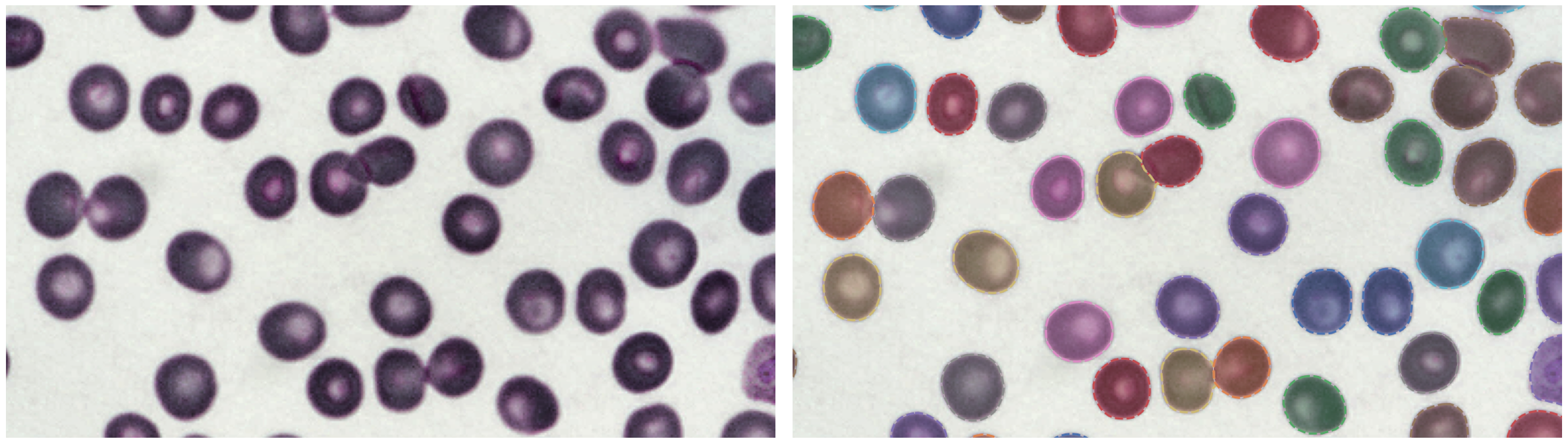
https://bbbc.broadinstitute.org/BBBC041 (CC BY-NC-SA 3.0)
🛠 Install
Make sure you have PyTorch installed.
PyPI
pip install -U celldetection
GitHub
pip install git+https://github.com/FZJ-INM1-BDA/celldetection.git
💾 Trained models
model = cd.fetch_model(model_name, check_hash=True)
| model name | training data | link |
|---|---|---|
ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c | BBBC039, BBBC038, Omnipose, Cellpose, Sartorius - Cell Instance Segmentation, Livecell, NeurIPS 22 CellSeg Challenge | 🔗 |
🐳 Docker
Find us on Docker Hub: https://hub.docker.com/r/ericup/celldetection
You can pull the latest version of celldetection via:
doker pull ericup/celldetection:latest
Apptainer
You can also pull our Docker images for the use with Apptainer (formerly Singularity) with this command:
apptainer pull --dir . --disable-cache docker://ericup/celldetection:latest
🤗 Hugging Face Spaces
Find us on Hugging Face and upload your own images for segmentation: https://huggingface.co/spaces/ericup/celldetection
🧑💻 Napari Plugin
Find our Napari Plugin here: https://github.com/FZJ-INM1-BDA/celldetection-napari
Find out more about Napari here: https://napari.org
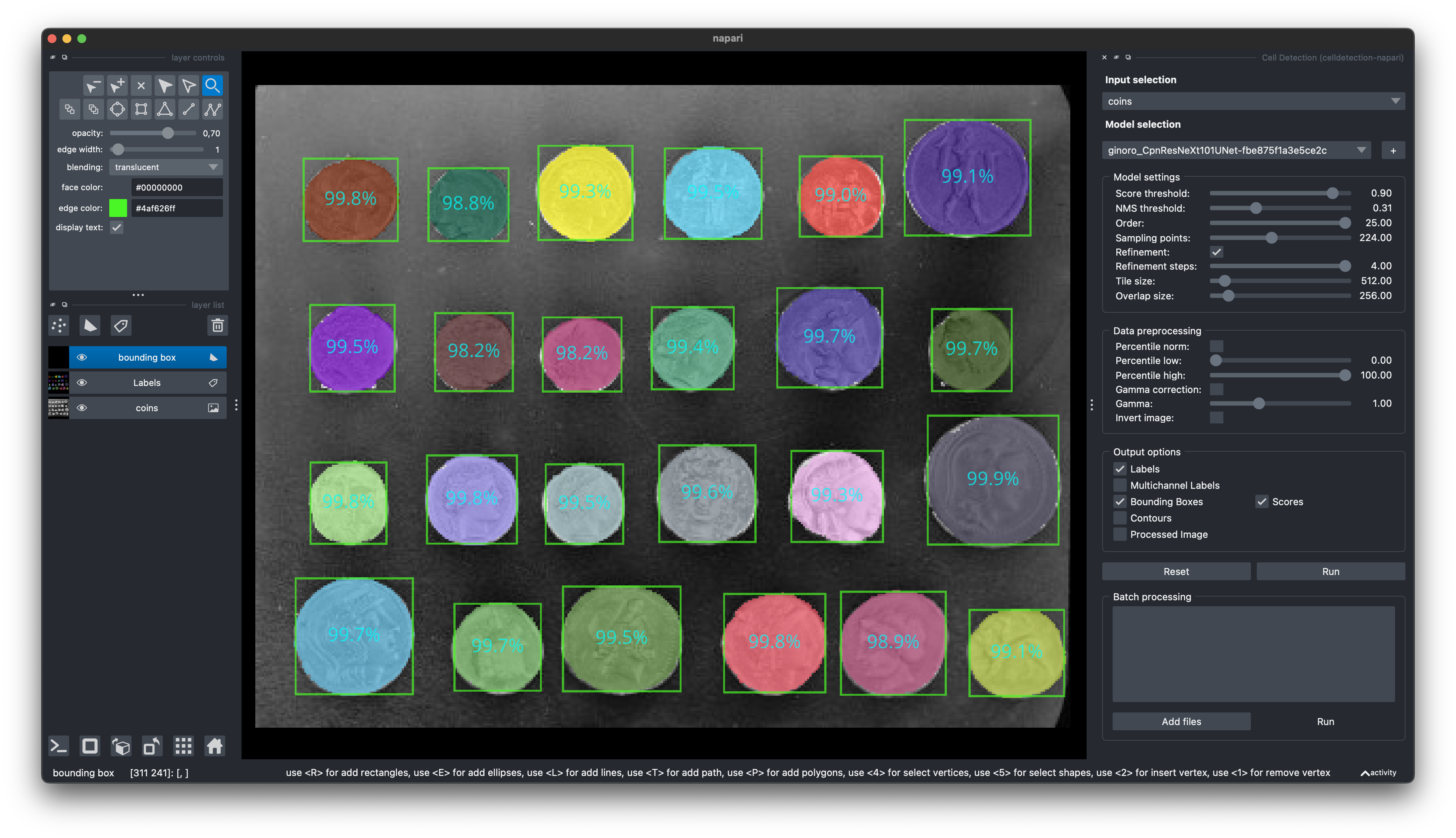
You can install it via pip:
pip install git+https://github.com/FZJ-INM1-BDA/celldetection-napari.git
🏆 Awards
- NeurIPS 2022 Cell Segmentation Challenge: Winner Finalist Award
📝 Citing
If you find this work useful, please consider giving a star ⭐️ and citation:
@article{UPSCHULTE2022102371,
title = {Contour proposal networks for biomedical instance segmentation},
journal = {Medical Image Analysis},
volume = {77},
pages = {102371},
year = {2022},
issn = {1361-8415},
doi = {https://doi.org/10.1016/j.media.2022.102371},
url = {https://www.sciencedirect.com/science/article/pii/S136184152200024X},
author = {Eric Upschulte and Stefan Harmeling and Katrin Amunts and Timo Dickscheid},
keywords = {Cell detection, Cell segmentation, Object detection, CPN},
}
🔗 Links

- Apache-2.0
Participating organisations
Reference papers
- 1.Author(s): Jun Ma, Ronald Xie, Shamini Ayyadhury, Cheng Ge, Anubha Gupta, Ritu Gupta, Song Gu, Yao Zhang, Gihun Lee, Joonkee Kim, Wei Lou, Haofeng Li, Eric Upschulte, Timo Dickscheid, José Guilherme de Almeida, Yixin Wang, Lin Han, Xin Yang, Marco Labagnara, Vojislav Gligorovski, Maxime Scheder, Sahand Jamal Rahi, Carly Kempster, Alice Pollitt, Leon Espinosa, Tâm Mignot, Jan Moritz Middeke, Jan-Niklas Eckardt, Wangkai Li, Zhaoyang Li, Xiaochen Cai, Bizhe Bai, Noah F. Greenwald, David Van Valen, Erin Weisbart, Beth A. Cimini, Trevor Cheung, Oscar Brück, Gary D. Bader, Bo WangPublished in Nature Methods by Springer Science and Business Media LLC in 2024, page: 1103-111310.1038/s41592-024-02233-6
- 2.Author(s): Eric Upschulte, Stefan Harmeling, Katrin Amunts, Timo DickscheidPublished in Medical Image Analysis by Elsevier BV in 2022, page: 10237110.1016/j.media.2022.102371
Mentions
- 1.Author(s): Timo Kaiser, Vladimír Ulman, Bodo RosenhahnPublished in Lecture Notes in Computer Science, Computer Vision – ECCV 2024 Workshops by Springer Nature Switzerland in 2025, page: 122-13810.1007/978-3-031-91721-9_8
- 2.Author(s): Junfeng Chen, Yuzhu Liu, Mengzhao Yao, Jingjing DuPublished in Communications in Computer and Information Science, Data Mining and Big Data by Springer Nature Singapore in 2025, page: 302-31210.1007/978-981-96-7175-5_25
- 3.Author(s): Zdravko Marinov, Alexander Jaus, Jens Kleesiek, Rainer StiefelhagenPublished in Lecture Notes in Computer Science, Medical Image Segmentation Foundation Models. CVPR 2024 Challenge: Segment Anything in Medical Images on Laptop by Springer Nature Switzerland in 2025, page: 39-5610.1007/978-3-031-81854-7_3
- 4.Author(s): L. Maddalena, L. Antonelli, F. Polverino, I. A. Asteriti, F. Degrassi, G. Guarguaglini, A. Albu, A. Hada, M. R. GuarracinoPublished in Trends in Biomathematics: Modeling Health Across Ecology, Social Interactions, and Cells by Springer Nature Switzerland in 2025, page: 199-21910.1007/978-3-031-97461-8_11
- 5.Author(s): Songxiao Yang, Yizhou Li, Ye Chen, Zhuofeng Wu, Masatoshi OkutomiPublished in Lecture Notes in Computer Science, Medical Image Segmentation Foundation Models. CVPR 2024 Challenge: Segment Anything in Medical Images on Laptop by Springer Nature Switzerland in 2025, page: 83-10010.1007/978-3-031-81854-7_6
- 6.Author(s): Johannes Seiffarth, Luisa Blöbaum, Richard D. Paul, Nils Friederich, Angelo Jovin Yamachui Sitcheu, Ralf Mikut, Hanno Scharr, Alexander Grünberger, Katharina NöhPublished in Lecture Notes in Computer Science, Computer Vision – ECCV 2024 Workshops by Springer Nature Switzerland in 2025, page: 318-33410.1007/978-3-031-91721-9_20
- 7.Author(s): Zdravko Marinov, Alexander Jaus, Jens Kleesiek, Rainer StiefelhagenPublished in Lecture Notes in Computer Science, Medical Image Segmentation Foundation Models. CVPR 2024 Challenge: Segment Anything in Medical Images on Laptop by Springer Nature Switzerland in 2025, page: 101-12510.1007/978-3-031-81854-7_7
- 8.Author(s): Youngbin Kong, Kwangtai Kim, Seoi Jeong, Kyu Eun Lee, Hyoun-Joong KongPublished in Lecture Notes in Computer Science, Medical Image Segmentation Foundation Models. CVPR 2024 Challenge: Segment Anything in Medical Images on Laptop by Springer Nature Switzerland in 2025, page: 180-19410.1007/978-3-031-81854-7_12
- 9.Author(s): Trung DQ. Dang, Huy Hoang Nguyen, Aleksei TiulpinPublished in Lecture Notes in Computer Science, Computer Vision – ACCV 2024 by Springer Nature Singapore in 2024, page: 222-24010.1007/978-981-96-0901-7_14
- 10.Author(s): Chaoyu Chen, Xin Yang, Rusi Chen, Junxuan Yu, Liwei Du, Jian Wang, Xindi Hu, Yan Cao, Yingying Liu, Dong NiPublished in Lecture Notes in Computer Science, Machine Learning in Medical Imaging by Springer Nature Switzerland in 2023, page: 166-17510.1007/978-3-031-45673-2_17
- 11.Author(s): S. J. van Albada, A. Morales-Gregorio, T. Dickscheid, A. Goulas, R. Bakker, S. Bludau, G. Palm, C.-C. Hilgetag, M. DiesmannPublished in Advances in Experimental Medicine and Biology, Computational Modelling of the Brain by Springer International Publishing in 2021, page: 201-23410.1007/978-3-030-89439-9_9
- 1.Author(s): Valentina Vadori, Antonella Peruffo, Jean-Marie Graïe, Livio Finos, Enrico GrisanPublished in 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI) by IEEE in 2025, page: 1-410.1109/isbi60581.2025.10980998
- 2.Author(s): Philipp Endres, Valentin Koch, Julia A. Schnabel, Carsten MarrPublished in 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI) by IEEE in 2025, page: 1-510.1109/isbi60581.2025.10980782
- 3.Author(s): Yuming Chen, Qi Wu, Yutong XiePublished in 2025 International Conference on Digital Image Computing: Techniques and Applications (DICTA) by IEEE in 2025, page: 1-810.1109/dicta68720.2025.11302485
- 4.Author(s): Manh Duong Nguyen, Nguyen Dang Huy Pham, Phi Le Nguyen, Minh N. DoPublished in 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI) by IEEE in 2025, page: 1-410.1109/isbi60581.2025.10981240
- 5.Author(s): Lingbo Zhang, Bingqian Sun, Linghan Cai, Yifeng Wang, Ye Zhang, Songhan Jiang, Kai Zhang, Yongbing ZhangPublished in Proceedings of the 33rd ACM International Conference on Multimedia by ACM in 2025, page: 2900-290810.1145/3746027.3754883
- 6.Author(s): Pawel Tomasz Pieta, Peter Winkel Rasmussen, Anders Bjorholm Dahl, Anders Nymark ChristensenPublished in 2025 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) by IEEE in 2025, page: 4667-467710.1109/cvprw67362.2025.00453
- 7.Author(s): Jan-Oliver Kropp, Christian Schiffer, Katrin Amunts, Timo DickscheidPublished in 2024 IEEE International Symposium on Biomedical Imaging (ISBI) by IEEE in 2024, page: 1-510.1109/isbi56570.2024.10635384
- 8.Author(s): Giulia Ciacci, Cristiano D'Andrea, Paolo Matteini, Elisa Maria Alessi, Sara Di Ruzza, Maria Antonietta Pascali, Sara Colantonio, Ambra Giannetti, Andrea BarucciPublished in 2024 Italian Conference on Optics and Photonics (ICOP) by IEEE in 2024, page: 1-410.1109/icop62013.2024.10803634
- 9.Author(s): Lihui Ding, Hongliang Wang, Lijun FuPublished in 2024 10th International Conference on Computer and Communications (ICCC) by IEEE in 2024, page: 781-78510.1109/iccc62609.2024.10942266
- 10.Author(s): Tianrun Chen, Chaotao Ding, Lanyun Zhu, Tao Xu, Yan Wang, Deyi Ji, Ying Zang, Zejian LiPublished in 2024 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI) by IEEE in 2024, page: 1-810.1109/bhi62660.2024.10913659
- 11.Author(s): Aleksei Samarin, Alexander Savelev, Aleksei ToropovPublished in 2024 International Russian Automation Conference (RusAutoCon) by IEEE in 2024, page: 331-33710.1109/rusautocon61949.2024.10694216
- 12.Author(s): Neel Dey, S. Mazdak Abulnaga, Benjamin Billot, Esra Abaci Turk, P. Ellen Grant, Adrian V. Dalca, Polina GollandPublished in 2024 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) by IEEE in 2024, page: 7578-758810.1109/wacv57701.2024.00742
- 13.Author(s): Futai Pan, Fei WangPublished in 2023 IEEE International Conference on Medical Artificial Intelligence (MedAI) by IEEE in 2023, page: 334-34310.1109/medai59581.2023.00052
- 14.Author(s): Eric Upschulte, Stefan Harmeling, Katrin Amunts, Timo DickscheidPublished in Competitions in Neural Information Processing Systems by PMLR in 2023, page: 1-12
- 15.Author(s): Qi Chen, Wei Huang, Xiaoyu Liu, Jiacheng Li, Zhiwei XiongPublished in 2023 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW) by IEEE in 2023, page: 3905-391410.1109/iccvw60793.2023.00422
- 1.Author(s): Xiaotong Wang, Rong Xiao, Hao Wei, Song Yang, Shangbin Chen, Yicheng XiePublished in Biomedical Signal Processing and Control by Elsevier BV in 2026, page: 10867710.1016/j.bspc.2025.108677
- 2.Author(s): Begüm Yetişkin, Selami Çalışkan, İsmail Güney, Çetin Erçelik, M. Fatih TaluPublished in Politeknik Dergisi by Politeknik Dergisi in 202610.2339/politeknik.1660457
- 3.Author(s): Hancan Zhu, Jinhao Chen, Guanghua HePublished in Biomedical Signal Processing and Control by Elsevier BV in 2026, page: 10882110.1016/j.bspc.2025.108821
- 4.Author(s): Eliotte E. Garling, Shally Li, Kristin Ho, Karine A. GibbsPublished in mBio by American Society for Microbiology in 202610.1128/mbio.03342-25
- 5.Author(s): Ziyao Zhang, Qiankun Ma, Tong Zhang, Jie Chen, Hairong Zheng, Wen GaoPublished in Medical Image Analysis by Elsevier BV in 2026, page: 10379210.1016/j.media.2025.103792
- 6.Author(s): Bogdan Kochetov, Shikhar UttamPublished in Computers in Biology and Medicine by Elsevier BV in 2025, page: 11029610.1016/j.compbiomed.2025.110296
- 7.Author(s): Zhen Chen, Qing Xu, Xinyu Liu, Yixuan YuanPublished in Medical Image Analysis by Elsevier BV in 2025, page: 10360710.1016/j.media.2025.103607
- 8.Author(s): Felix Y. Zhou, Zach Marin, Clarence Yapp, Qiongjing Zou, Benjamin A. Nanes, Stephan Daetwyler, Andrew R. Jamieson, Md Torikul Islam, Edward Jenkins, Gabriel M. Gihana, Jinlong Lin, Hazel M. Borges, Bo-Jui Chang, Andrew Weems, Sean J. Morrison, Peter K. Sorger, Reto Fiolka, Kevin M. Dean, Gaudenz DanuserPublished in Nature Methods by Springer Science and Business Media LLC in 2025, page: 2386-239910.1038/s41592-025-02887-w
- 9.Author(s): Federica Furlanetto, Nicole Flegel, Marco Kremp, Chiara Spear, Franziska Fröb, Margherita Alfonsetti, Bettina Bohl, Mandy Krumbiegel, Sören Turan, Andre Reis, Dieter C Lie, Jürgen Winkler, Sven Falk, Michael Wegner, Marisa KarowPublished in Life Science Alliance by Life Science Alliance, LLC in 2025, page: e20240310210.26508/lsa.202403102
- 10.Author(s): Henrique Quaiato de Oliveira, Solon Andrades da Rosa, Luiza Cherobini Pereira, Laura Boose de Mendonça, Juliete Scholl, Fabrício Figueiró, Thais Cardoso Bitencourt, Davi Piovesan Echevarria, João Luiz Aldinucci Buzzo, Jessica Boschini D'Agostin, Danieli Rosane Dallemole, Carolina Oliveira, Fernanda Simas, Fernanda Saez‐Calazans, Débora Santos‐Sousa, Guido Lenz, Eduardo C. Filippi‐ChielaPublished in The FEBS Journal by Wiley in 202510.1111/febs.70339
- 11.Author(s): Depeng Gao, Ying Gong, Jingzhuo Cao, Bingshu Wang, Han Zhang, Jiangkai Dong, Jianlin QiuPublished in Electronics by MDPI AG in 2025, page: 47910.3390/electronics14030479
- 12.Author(s): Takaaki Aratake, Serika Kurita, Michael WegnerPublished in Cells by MDPI AG in 2025, page: 175610.3390/cells14221756
- 13.Author(s): Yichen Si, Joo Sang Lee, Goo Jun, Hyun Min Kang, Jun Hee LeePublished in Trends in Genetics by Elsevier BV in 2025, page: 774-78710.1016/j.tig.2025.05.002
- 14.Author(s): Rui Ma, Jia Qian, Xing Li, Siying Wang, Chen Bai, Xianghua Yu, Taiqiang Dai, Liang Kong, Dan Dan, Baoli YaoPublished in Optics and Lasers in Engineering by Elsevier BV in 2025, page: 10895010.1016/j.optlaseng.2025.108950
- 15.Author(s): Matthias Bruhns, Jan T. Schleicher, Maximilian Wirth, Marcello Zago, Sepideh Babaei, Manfred ClaassenPublished in PLOS Computational Biology by Public Library of Science (PLoS) in 2025, page: e101335010.1371/journal.pcbi.1013350
- 16.Author(s): Jan Clusmann, Maria Balaguer-Montero, Octavi Bassegoda, Carolin V. Schneider, Tobias Seraphin, Ellis Paintsil, Tom Luedde, Raquel Perez Lopez, Julien Calderaro, Stephen Gilbert, Thomas Marjot, Ashley Spann, Debbie L. Shawcross, Sabela Lens, Eric Trépo, Jakob Nikolas KatherPublished in Journal of Hepatology by Elsevier BV in 2025, page: 1410-142610.1016/j.jhep.2025.07.003
- 17.Author(s): Jintu Zheng, Qizhe Liu, Yi Ding, Yi Cao, Ying Hu, Zenan WangPublished in Medical Image Analysis by Elsevier BV in 2025, page: 10367510.1016/j.media.2025.103675
- 18.Author(s): Zhaoke Huang, Zelin Li, Hong YanPublished in IEEE Transactions on Computational Biology and Bioinformatics by Institute of Electrical and Electronics Engineers (IEEE) in 2025, page: 890-89810.1109/tcbbio.2025.3542123
- 19.Author(s): Qinqin Huang, Shen’ao Wu, Zhenkai Ou, Yi GaoPublished in Intelligent Oncology by Elsevier BV in 2025, page: 139-15910.1016/j.intonc.2025.03.004
- 20.Author(s): Can Shi, Jinghong Fan, Zhonghan Deng, Huanlin Liu, Qiang Kang, Yumei Li, Jing Guo, Jingwen Wang, Jinjiang Gong, Sha Liao, Ao Chen, Ying Zhang, Mei LiPublished in GigaScience by Oxford University Press (OUP) in 202510.1093/gigascience/giaf069
- 21.Author(s): Jhonatan Salgado, James Rayner, Nikola OjkicPublished in Frontiers in Microbiology by Frontiers Media SA in 202510.3389/fmicb.2025.1536131
- 22.Author(s): Carsen Stringer, Marius PachitariuPublished in Nature Methods by Springer Science and Business Media LLC in 2025, page: 592-59910.1038/s41592-025-02595-5
- 23.Author(s): Lidan Wu, Joseph M. Beechem, Patrick DanaherPublished in Scientific Reports by Springer Science and Business Media LLC in 202510.1038/s41598-025-08733-5
- 24.Author(s): Xiao Li, Qinxue Ouyang, Mingfei Han, Xiaoqing Liu, Fuchu He, Yunping Zhu, Ling Leng, Jie MaPublished in Advanced Intelligent Systems by Wiley in 202510.1002/aisy.202400635
- 25.Author(s): Abishek Sankaranarayanan, Georgii Khachaturov, Kimberly S. Smythe, Shachi MittalPublished in Communications Biology by Springer Science and Business Media LLC in 202510.1038/s42003-025-08184-8
- 26.Author(s): Jun Hu, Zhiwen Liao, Chengyan Bian, Di Gai, Xingchen Wu, Pengxiang Su, Yueqiu Qin, Yuan Ni, Zhiruo Zhang, Hengrui Liu, Qi Wang, Hua LiPublished in Alexandria Engineering Journal by Elsevier BV in 2025, page: 1029-104310.1016/j.aej.2025.09.045
- 27.Author(s): Ameer Sarwar, Mara Rue, Leon French, Helen Cross, Sarah Choi, Xiaoyin Chen, Jesse GillisPublished in Genome Biology by Springer Science and Business Media LLC in 202510.1186/s13059-025-03747-8
- 28.Author(s): Ilya Larin, Alexander KarabelskyPublished in Journal of Imaging by MDPI AG in 2025, page: 10310.3390/jimaging11040103
- 29.Author(s): Ching-Wei Wang, Kai-Lin Chu, Ting-Sheng Su, Keng-Wei Liu, Yi-Jia Lin, Tai-Kuang ChaoPublished in IEEE Journal of Biomedical and Health Informatics by Institute of Electrical and Electronics Engineers (IEEE) in 2025, page: 333-34410.1109/jbhi.2024.3476554
- 30.Author(s): Shu-Yu Guo, Lan Huang, Yu-Hao Mu, Tian BaiPublished in Journal of Computer Science and Technology by Springer Science and Business Media LLC in 2025, page: 780-79110.1007/s11390-025-4324-4
- 31.Author(s): Markus Marks, Uriah Israel, Rohit Dilip, Qilin Li, Changhua Yu, Emily Laubscher, Ahamed Iqbal, Elora Pradhan, Ada Ates, Martin Abt, Caitlin Brown, Edward Pao, Shenyi Li, Alexander Pearson-Goulart, Pietro Perona, Georgia Gkioxari, Ross Barnowski, Yisong Yue, David Van ValenPublished in Nature Methods by Springer Science and Business Media LLC in 2025, page: 2585-259310.1038/s41592-025-02879-w
- 32.Author(s): Cyril Achard, Timokleia Kousi, Markus Frey, Maxime Vidal, Yves Paychere, Colin Hofmann, Asim Iqbal, Sebastien B Hausmann, Stéphane Pagès, Mackenzie Weygandt MathisPublished in eLife by eLife Sciences Publications, Ltd in 202510.7554/elife.99848
- 33.Author(s): Ching-Wei Wang, Wei-Tang Lee, Ting-Sheng SuPublished in Neural Computing and Applications by Springer Science and Business Media LLC in 2025, page: 11195-1126410.1007/s00521-025-11119-3
- 34.Author(s): Chun Kiet Vong, Alan Wang, Mike Dragunow, Thomas I-H. Park, Vickie ShimPublished in Computers in Biology and Medicine by Elsevier BV in 2025, page: 10964210.1016/j.compbiomed.2024.109642
- 35.Author(s): Ziwen Liu, Eduardo Hirata-Miyasaki, Soorya Pradeep, Johanna V. Rahm, Christian Foley, Talon Chandler, Ivan E. Ivanov, Hunter O. Woosley, See-Chi Lee, Sudip Khadka, Tiger Lao, Akilandeswari Balasubramanian, Rita Marreiros, Chad Liu, Camille Januel, Manuel D. Leonetti, Ranen Aviner, Carolina Arias, Adrian Jacobo, Shalin B. MehtaPublished in Nature Machine Intelligence by Springer Science and Business Media LLC in 2025, page: 901-91510.1038/s42256-025-01046-2
- 36.Author(s): Anwai Archit, Luca Freckmann, Sushmita Nair, Nabeel Khalid, Paul Hilt, Vikas Rajashekar, Marei Freitag, Carolin Teuber, Melanie Spitzner, Constanza Tapia Contreras, Genevieve Buckley, Sebastian von Haaren, Sagnik Gupta, Marian Grade, Matthias Wirth, Günter Schneider, Andreas Dengel, Sheraz Ahmed, Constantin PapePublished in Nature Methods by Springer Science and Business Media LLC in 2025, page: 579-59110.1038/s41592-024-02580-4
- 37.Author(s): Jordão Bragantini, Ilan Theodoro, Xiang Zhao, Teun A.P.M. Huijben, Eduardo Hirata-Miyasaki, Shruthi VijayKumar, Akilandeswari Balasubramanian, Tiger Lao, Richa Agrawal, Sheng Xiao, Jan Lammerding, Shalin Mehta, Alexandre X. Falcão, Adrian Jacobo, Merlin Lange, Loïc A. RoyerPublished in Nature Methods by Springer Science and Business Media LLC in 2025, page: 2423-243610.1038/s41592-025-02778-0
- 38.Author(s): Johannes Seiffarth, Katharina NöhPublished in PLOS One by Public Library of Science (PLoS) in 2025, page: e033711010.1371/journal.pone.0337110
- 39.Author(s): Renpeng Ding, Kerem Celikay, Ming Ni, Yong Hou, Yan Zhou, Karl RohrPublished in Small Methods by Wiley in 202510.1002/smtd.202500885
- 40.Author(s): Siyuan Dai, Kai Ye, Charlie Zhan, Haoteng Tang, Liang ZhanPublished in Computational and Structural Biotechnology Journal by Elsevier BV in 2025, page: 744-75210.1016/j.csbj.2025.02.024
- 41.Author(s): Patcharaporn Boottanun, Sayaka Fuseya, Atsushi KunoPublished in BBA Advances by Elsevier BV in 2025, page: 10014610.1016/j.bbadva.2025.100146
- 42.Author(s): Ruoyun Liu, Jianshu Chao, Jiahua Lai, Qingwei Guo, Ke Sun, Zeyu ZhangPublished in The Visual Computer by Springer Science and Business Media LLC in 202510.1007/s00371-025-04211-1
- 43.Author(s): Joshua Millward, Zhen He, Aiden Nibali, Dmitri Mouradov, Lisa A Mielke, Kelly Tran, Angela Chou, Nicholas J Hawkins, Robyn L Ward, Anthony J Gill, Oliver M Sieber, David S WilliamsPublished in Journal of Translational Medicine by Springer Science and Business Media LLC in 202510.1186/s12967-025-06254-3
- 44.Author(s): Chong Wang, Yajie Wan, Shuxin Li, Kaili Qu, Xuezhi Zhou, Junjun He, Jing Ke, Yi Yu, Tianyun Wang, Yiqing ShenPublished in IEEE Transactions on Medical Imaging by Institute of Electrical and Electronics Engineers (IEEE) in 2025, page: 3924-393710.1109/tmi.2024.3501352
- 45.Author(s): Gabriel Casella, Madeleine S. Torcasso, Junting Ai, Thao P. Cao, Satoshi Hara, Michael S. Andrade, Deepjyoti Ghosh, Daming Shao, Anthony Chang, Kichul Ko, Anita S. Chong, Maryellen L. Giger, Marcus R. ClarkPublished in Journal of Clinical Investigation by American Society for Clinical Investigation in 202510.1172/jci192669
- 46.Author(s): Cyril Achard, Timokleia Kousi, Markus Frey, Maxime Vidal, Yves Paychere, Colin Hofmann, Asim Iqbal, Sebastien B Hausmann, Stéphane Pagès, Mackenzie Weygandt MathisPublished in eLife by eLife Sciences Publications, Ltd in 202510.7554/elife.99848.4
- 47.Author(s): Shizhe Sun, Wataru OhyamaPublished in SN Computer Science by Springer Science and Business Media LLC in 202510.1007/s42979-025-04267-9
- 48.Author(s): Tim Van De Looverbosch, Sarah De Beuckeleer, Frederik De Smet, Jan Sijbers, Winnok H. De VosPublished in Computers in Biology and Medicine by Elsevier BV in 2025, page: 10956110.1016/j.compbiomed.2024.109561
- 49.Author(s): Quan Sun, Lei Wang, Jun Ma, Fei He, Dongyu Wei, Chunming SiPublished in Frontiers in Medicine by Frontiers Media SA in 202510.3389/fmed.2025.1615699
- 50.Author(s): Valentina Vadori, Jean-Marie Graïc, Antonella Peruffo, Giulia Vadori, Livio Finos, Enrico GrisanPublished in Computers in Biology and Medicine by Elsevier BV in 2025, page: 11101810.1016/j.compbiomed.2025.111018
- 1.Author(s): Omid Sharafi, Timothy Silliman, Hadi Mokhtari Dowlatabad, Gengle Niu, Steven T. Walston, Jean-Marie C. Bouteiller, Kimberly K. Gokoffski, Gianluca LazziPublished in 202610.64898/2026.01.22.701180
- 2.Author(s): John Hageter, Audrey DelGaudio, Maegan Leathery, Braxton Johnson, Tegan Raupp, James Holcomb, Axel Faz Treviño, Julius Jonaitis, Morgan S. Bridi, Andrew Dacks, Eric J. HorstickPublished in 202610.1016/j.crmeth.2025.101298
- 3.Author(s): Thomas Defard, Alice Blondel, Sebastien Bellow, Anthony Coleon, Guilherme Dias de Melo, Florian Mueller, Thomas WalterPublished in 202610.1186/s13059-025-03908-9
- 4.Author(s): Mario Koddenbrock, Justus Westerhoff, Dominik Fachet, Simone Reber, Felix Gers, Erik RodnerPublished in 202610.64898/2026.01.09.698597
- 5.Author(s): Guoqing Zhang, Shichao Kan, Yigang Cen, Yigang Cen, Yi Cen, Yi Cen, Qi Cao, Yansen Huang, Ming ZengPublished in 202610.1016/j.jvcir.2026.104739
- 6.Author(s): Katariina Luukkainen, Mikko Purhonen, Mikael Tatun, Kevin Hung, Oda Tafjord, Henri Sundquist, Stina Söderlund, Shady Adnan‐Awad, Anni Dohlen, Johanna Heikkinen, Perttu Koskenvesa, Lotta Joutsi‐Korhonen, Anna Lempiäinen, Sanna Siitonen, Satu Mustjoki, Naranie Shanmuganathan, Coral Bryce, Signe Danielsson, Henrik Hjorth‐Hansen, Ulla Olsson‐Strömberg, Takashi Kumagai, Shinya Kimura, David M. Ross, Oscar BrückPublished in 202610.1002/hem3.70319
- 7.Author(s): Dilan Martínez-Torres, Nicolás Bettancourt, Jorge Vergara-Estrada, Karen Almendras-Durán, Valeria Candia, Fabián Segovia-Miranda, Hernán Morales-NavarretePublished in 202610.21203/rs.3.rs-8903621/v1
- 8.Author(s): Yiwen Li, Yusheng Zhang, Ying Zhang, Qing Wang, Boyang Ji, Hongjun Yang, Xianyu LiPublished in 202610.1093/pcmedi/pbaf040
- 9.Author(s): Peilin Zhou, Jiongyi Wang, Yanmin Dai, Hanyu Du, Yuliang Gu, Yongchao XuPublished in 202610.1016/j.neucom.2026.132890
- 10.Author(s): Deboch Eyob Abera, Jiaye He, Jia Liu, Nazar Zaki, Wenjian QinPublished in 202610.1016/j.displa.2025.103337
- 11.Author(s): Mayuri Vaish, Benjamin A. NanesPublished in 202610.1016/j.jid.2026.01.007
- 12.Author(s): Jinhao Chen, Chao Ma, Jinping Chen, Zhong Li, Zhaoxi Fang, Keli Hu, Guanghua He, Hancan ZhuPublished by Springer Science and Business Media LLC in 202510.21203/rs.3.rs-8244943/v1
- 13.Author(s): Naiqiao Hou, Yue Yu, Zhaorun Wu, Yanqi Zhang, Jiayun Wu, Zhixing Zhong, Fangfang Xu, Zeyu Wang, Chaoyong Yang, Weihong Tan, Jia SongPublished by openRxiv in 202510.1101/2025.04.07.647590
- 14.Author(s): Thomas Defard, Alice Blondel, Sebastien Bellow, Anthony Coleon, Guilherme Dias de Melo, Thomas Walter, Florian MuellerPublished by openRxiv in 202510.1101/2025.03.03.641259
- 15.Author(s): Rémy Torro, Beatriz Díaz-Bello, Dalia El Arawi, Ksenija Dervanova, Lorna Ammer, Florian Dupuy, Patrick Chames, Kheya Sengupta, Laurent LimozinPublished by eLife Sciences Publications, Ltd in 202510.7554/elife.105302.1
- 16.Author(s): Gabriel Casella, Madeleine Torcasso, Junting Ai, Thao Cao, Satoshi Hara, Michael Andrade, Deepjyoti Ghosh, Anthony Chang, Kichul Ko, Anita S. Chong, Maryellen Giger, Marcus R. ClarkPublished by openRxiv in 202510.1101/2025.02.17.638655
- 17.Author(s): Federica Furlanetto, Alejandro Segura, Mathar Kravikass, Pritha Dolai, Sarah Frank, Sören Turan, Angelica Luna Leal, Stephan Käseberg, Susann Schweiger, Chichung D. Lie, Vasily Zaburdaev, Marisa Karow, Sven FalkPublished by openRxiv in 202510.1101/2025.04.15.648981
- 18.Author(s): Xiao Liu, Chenxu Zhang, Fuxiang Huang, Shuyin Xia, Guoyin Wang, Lei ZhangPublished in 202510.1109/tnnls.2025.3610435
- 19.Author(s): Lukas Arnroth, Sanja VickovicPublished by openRxiv in 202510.1101/2025.05.02.651831
- 20.Author(s): Marius Pachitariu, Michael Rariden, Carsen StringerPublished by openRxiv in 202510.1101/2025.04.28.651001
- 21.Author(s): Shuo Zhao, Jianxu ChenPublished in 202510.1109/bibm66473.2025.11356754
- 22.Author(s): John Hageter, Audrey DelGaudio, Maegan Leathery, Braxton Johnson, Tegan Raupp, James Holcomb, Axel Faz Treviño, Julius Jonaitis, Morgan S. Bridi, Andrew Dacks, Eric J. HorstickPublished by openRxiv in 202510.1101/2025.08.19.671108
- 23.Author(s): Can Ergen, Nir YosefPublished by openRxiv in 202510.1101/2025.01.20.634005
- 24.Author(s): Rémy Torro, Beatriz Díaz-Bello, Dalia El Arawi, Ksenija Dervanova, Lorna Ammer, Florian Dupuy, Patrick Chames, Kheya Sengupta, Laurent LimozinPublished by eLife Sciences Publications, Ltd in 202510.7554/elife.105302
- 25.Author(s): Can Shi, Yumei Li, Jing Guo, Qiuling Chen, Tingting Cao, Sha Liao, Ao Chen, Mei Li, Ying ZhangPublished in 202510.1093/bib/bbaf718
- 26.Author(s): Can Shi, Yumei Li, Jing Guo, Qiuling Chen, Tingting Cao, Sha Liao, Ao Chen, Mei Li, Ying ZhangPublished by openRxiv in 202510.1101/2025.09.14.675731
- 27.Author(s): Wei Lou, Haofeng Li, Guanbin Li, Xiaoying Lou, Yuanhuan Xiong, Xiang Wan, Xusheng WuPublished in 202510.1109/tmi.2025.3583014
- 28.Author(s): Elyas Heidari, Andrew Moorman, Dániel Unyi, Nikhita Pasnuri, Gleb Rukhovich, Domenico Calafato, Anna Mathioudaki, Joseph M. Chan, Tal Nawy, Moritz Gerstung, Dana Pe’er, Oliver SteglePublished by openRxiv in 202510.1101/2025.03.14.643160
- 29.Author(s): Cyril Achard, Timokleia Kousi, Markus Frey, Maxime Vidal, Yves Paychère, Colin Hofmann, Asim Iqbal, Sebastien B Hausmann, Stéphane Pagès, Mackenzie Weygandt MathisPublished by eLife Sciences Publications, Ltd in 202510.7554/elife.99848.3
- 30.Author(s): Bogdan Kochetov, Shikhar UttamPublished by openRxiv in 202410.1101/2024.09.25.614955
- 31.Author(s): Xinsheng Wang, Xiaohua Wan, Buqing Cai, Zhuo Jia, Yuanbo Chen, Shuai Guo, Zheng Liu, Fuwei Li, Fa Zhang, Bin HuPublished by openRxiv in 202410.1101/2024.06.28.601295
- 32.Author(s): Jhonatan Salgado, James Rayner, Nikola OjkicPublished by MDPI AG in 202410.20944/preprints202408.2249.v1
- 33.Author(s): Cyril Achard, Timokleia Kousi, Markus Frey, Maxime Vidal, Yves Paychère, Colin Hofmann, Asim Iqbal, Sebastien B Hausmann, Stéphane Pagès, Mackenzie Weygandt MathisPublished by eLife Sciences Publications, Ltd in 202410.7554/elife.99848.1
- 34.Author(s): Rémy Torro, Beatriz Díaz-Bello, Dalia El Arawi, Ksenija Dervanova, Lorna Ammer, Florian Dupuy, Patrick Chames, Kheya Sengupta, Laurent LimozinPublished by openRxiv in 202410.1101/2024.03.15.585250
- 35.Author(s): Cyril Achard, Timokleia Kousi, Markus Frey, Maxime Vidal, Yves Paychère, Colin Hofmann, Asim Iqbal, Sebastien B Hausmann, Stéphane Pagès, Mackenzie Weygandt MathisPublished by eLife Sciences Publications, Ltd in 202410.7554/elife.99848.2
- 36.Author(s): Jordao Bragantini, Ilan Theodoro, Xiang Zhao, Teun A.P.M. Huijben, Eduardo Hirata-Miyasaki, Shruthi VijayKumar, Akilandeswari Balasubramanian, Tiger Lao, Richa Agrawal, Sheng Xiao, Jan Lammerding, Shalin Mehta, Alexandre Falcao, Adrian Jacobo, Merlin Lange, Loic A. RoyerPublished by openRxiv in 202410.1101/2024.09.02.610652
- 37.Author(s): Lidan Wu, Joseph M. Beechem, Patrick DanaherPublished by openRxiv in 202410.1101/2024.12.05.627051
- 38.Author(s): Ziwen Liu, Eduardo Hirata-Miyasaki, Soorya Pradeep, Johanna Rahm, Christian Foley, Talon Chandler, Ivan Ivanov, Hunter Woosley, Tiger Lao, Akilandeswari Balasubramanian, Rita Marreiros, Chad Liu, Manu Leonetti, Ranen Aviner, Carolina Arias, Adrian Jacobo, Shalin B. MehtaPublished by openRxiv in 202410.1101/2024.05.31.596901
- 39.Author(s): Saber Saharkhiz, Wesley Chan, Chloe B. Kirezi, Mearhyn Petite, Tony Roenspies, Theodore J. Perkins, Antonio ColavitaPublished by openRxiv in 202410.1101/2024.11.16.623955
- 40.Author(s): Julia Bonnet, Youssef El-Habouz, Célia Martin, Maelle Guillout, Louis Ruel, Baptiste Giroux, Claire Demeautis, Benjamin Mercat, Otmane Bouchareb, Jacques Pécreaux, Marc TramierPublished by openRxiv in 202410.1101/2024.09.24.614735
- 41.Author(s): Tim Van De Looverbosch, Sarah De Beuckeleer, Frederik De Smet, Jan Sijbers, Winnok H. De VosPublished by openRxiv in 202410.1101/2024.07.18.602066
- 42.Author(s): Zhiwen Hu, HaiBo Hong, Xuqiang Cai, Linxiang Li, Zichu Ren, Xi-Ao Ma, Haihua Jiang, Xun WangPublished by Springer Science and Business Media LLC in 202410.21203/rs.3.rs-5208094/v1
- 43.Author(s): Cyril Achard, Timokleia Kousi, Markus Frey, Maxime Vidal, Yves Paychère, Colin Hofmann, Asim Iqbal, Sebastien B. Hausmann, Stéphane Pagès, Mackenzie Weygandt MathisPublished by openRxiv in 202410.1101/2024.05.17.594691
- 44.Author(s): Can Shi, Jinghong Fan, Zhonghan Deng, Huanlin Liu, Qiang Kang, Yumei Li, Jing Guo, Jingwen Wang, Jinjiang Gong, Sha Liao, Ao Chen, Ying Zhang, Mei LiPublished by openRxiv in 202410.1101/2024.11.20.619750
- 45.Author(s): Carsen Stringer, Marius PachitariuPublished by openRxiv in 202410.1101/2024.04.06.587952
- 46.Author(s): Jun Ma, Ronald Xie, Shamini Ayyadhury, Cheng Ge, Anubha Gupta, Ritu Gupta, Song Gu, Yao Zhang, Gihun Lee, Joonkee Kim, Wei Lou, Haofeng Li, Eric Upschulte, Timo Dickscheid, José Guilherme de Almeida, Yixin Wang, Lin Han, Xin Yang, Marco Labagnara, Vojislav GligorovskiPublished by arXiv in 202310.48550/arxiv.2308.05864
- 47.Author(s): Amin Saberi, Casey Paquola, Konrad Wagstyl, Meike D. Hettwer, Boris C. Bernhardt, Simon B. Eickhoff, Sofie L. ValkPublished by openRxiv in 202310.1101/2023.03.25.532115
- 48.Author(s): Fei Liu, Zhixia Dong, Pei Qin, Binjie Qin, Sijie Xu, Xiangyun Zhao, Xinjian Wan, Xu Chen, Lu ChenPublished by Institute of Electrical and Electronics Engineers (IEEE) in 202310.36227/techrxiv.22186456.v3
- 49.Author(s): Uriah Israel, Markus Marks, Rohit Dilip, Qilin Li, Changhua Yu, Emily Laubscher, Ahamed Iqbal, Elora Pradhan, Ada Ates, Martin Abt, Caitlin Brown, Edward Pao, Shenyi Li, Alexander Pearson-Goulart, Pietro Perona, Georgia Gkioxari, Ross Barnowski, Yisong Yue, David Van ValenPublished by openRxiv in 202310.1101/2023.11.17.567630
- 50.Author(s): André Colliard-Granero, Mariah Batool, Jasna Jankovic, Jenia Jitsev, Michael H. Eikerling, Kourosh Malek, Mohammad Javad EslamibidgoliPublished by American Chemical Society (ACS) in 202110.26434/chemrxiv-2021-hr5zb

