FastSurfer
FastSurfer is a fast and accurate deep-learning pipeline for the analysis of human brain MRI. FastSurfer provides a fully compatible FreeSurfer alternative for volumetric and surface-based thickness analysis, also supporting sub-mm resolutions, and sub-segmentation of neuroanatomical structures.
Cite this software
Description
Quick Tutorial: Full Tutorial:
Full Documentation:

FastSurfer is a fast and accurate deep-learning based neuroimaging pipeline for the automated processing of structural human brain MRIs. Based on a T1-weighted MRI you get everything you need for quick structure localization (whole brain segmentation in under 1 minute), extraction of quantitative measures, group analysis of your cohort, or structural pre-processing for fMRI/diffusion analysis:
- Deep-learning based whole brain segmentation into 95 classes in under 1 minute on the GPU, and 20 min on the CPU
- Desikian-Killiany-Tourville atlas segmentation (=33 subcortical structures and 31 cortical structures / hemisphere)
- Cerebellar lobe segmentation into 27 structures
- ROI-wise volume statistics
- Supports native high-resolution segmentation (< 1.0mm^3, no resampling)
- Full FreeSurfer-conform outputs in approximately 45 min (+ 30 min for spherical registration)
- Cortical surfaces (white, pial)
- Surface measures (thickness, curvature and more)
- Surface labels and annotations (aparc.annot, cortex.label, ...)
- Point-wise surface statistics and ROI-wise volume statistics
- Spherical registration to FSAVERAGE - directly usable for inter-subject registration and alternative atlas registration (e.g. Destrieux, Desikian-Killiany)
- Supports native high-resolution MRIs (< 1.0mm^3, no resampling)
Check-out our Fastsurfer Colab Tutorial for a first taste of FastSurfer and generate your first FastSurfer segmentation in just three clicks!
Beyond the speed-up compared to traditional tools, FastSurfer and its components are highly accurate, reliable and sensitive to group effects. To ensure usefulness, we extensively validated the entire pipeline using a number of publicly available datasets (see Proof of Concept).
Further, running FastSurfer is easy as pie - a single command does all the work for you. The full documentation can be found on our github page.
./run_fastsurfer.sh \
--sd /home/user/my_fastsurfer_analysis \ # output dir (=SUBJECTS_DIR in FreeSurfer)
--sid SUBJECT_ID \ # result folder (auto-generated in output dir)
--t1 /home/user/my_mri_data/T1.nii.gz # Location of your T1-weighted MRI
We also provide Docker and Singularity container with all dependencies installed in it. You only need a valid FreeSurfer license for the surface stream, which is normally called .license or licence.txt and can be found in your FREESURFER_HOME directory or you can download it for free at https://surfer.nmr.mgh.harvard.edu/registration.html.
docker run --gpus all \
-v /home/user/my_mri_data:/DATA_INPUT_DIRECTORY \
-v /home/user/my_fastsurfer_analysis:/DATA_OUTPUT_DIRECTORY \
-v /home/user/my_fs_license_dir:/fs_license \
--rm --user $(id -u):$(id -g) \
deepmi/fastsurfer:latest
--sd DATA_OUTPUT_DIRECTORY \
--sid SUBJECT_ID \
--t1 DATA_INPUT_DIRECTORY/T1.nii.gz \
--fs_license /fs_license/.license \
Proof of Concept
1. Accuracy and Generalizability - How good is the whole brain segmentation generated by FastSurfer?
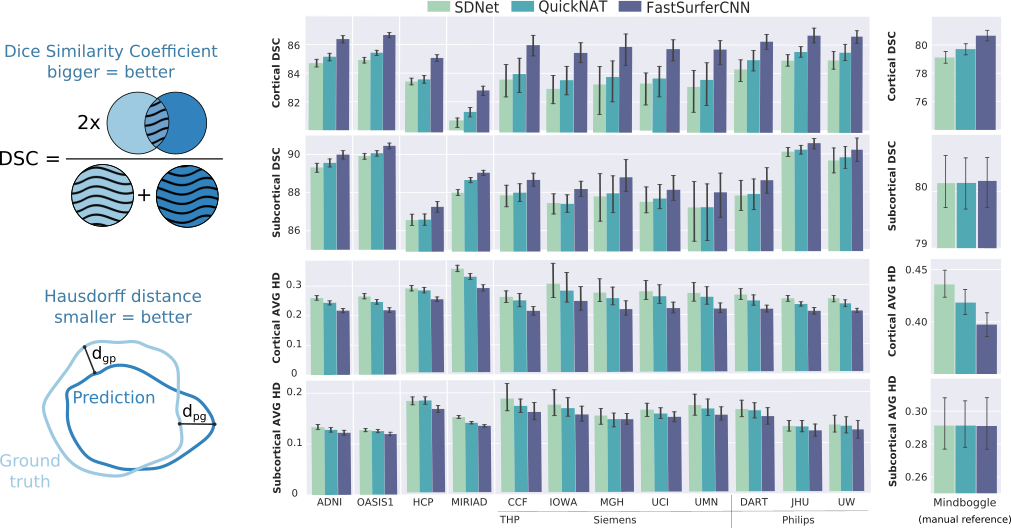
Answer: Very good! FastSurfer outperforms other deep-learning architectures by a significant margin both with respect to FreeSurfer and a manual standard (Mindboggle). It generalizes well across five different datasets including subjects with different disease states (e.g. cognitive normal, mild cognitive impaired or demented subjects in OASIS, ADNI, MIRIAD), different vendors (e.g. THP), age groups, downsampled and defaced images (e.g. HCP) and alternative T1-imaging protocols (e.g. MIRIAD)
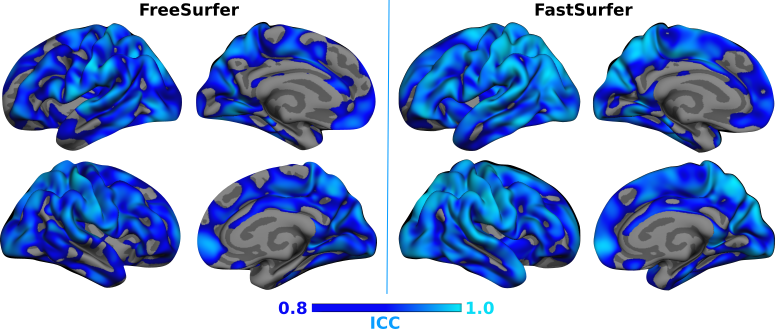
2. Reliability - How well does FastSurfer reproduce results across scans with minimal anatomical variations (Test-Retest)?
Answer: FastSurfer is highly reliable as demonstrated by the close agreement between the thickness and volumetric measurements for 20 Test-Retest subjects from OASIS1. FastSurfer exhibits high test-retest reliability and improves agreement between the cortical thickness measurements compared to FreeSurfer.
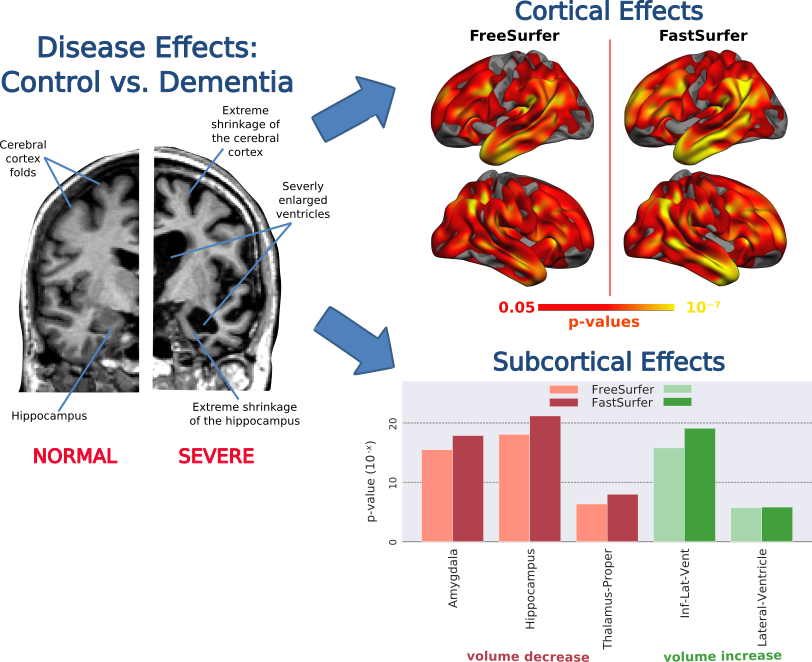
3. Sensitivity - How well does FastSurfer detect significant effects in the data?
Answer: FastSurfer is capable of accurately reproducing known disease effects in a control versus dementia cross-sectional group study. Reduced cortical thickness in regions associated with dementia (e.g. temporal lobes) as well as subcortical volume differences (e.g. enlarged ventricles, shrunken Hippocampus) are robustly detected with increased sensitivity relative to FreeSurfer.
Tool and Paper
-
FastSurfer is available as open source at github.
-
An accompanying set of quality control tools can be found at github.
-
In-depth information about FastSurfer can be found in our papers: FastSurfer, FastSurferVINN, CerebNet.
-
Or watch our small presentation (10min) on YouTube.
Tutorial
- A set of detailed tutorials for running Fastsurfer can be found within the FastSurfer github repository in the Tutorial Directory

- Apache-2.0
Participating organisations
Mentions
- 1.Author(s): Sol A. Cataldo, Florencia Sarmiento Laspiur, Martín A. BelzuncePublished in IFMBE Proceedings, Advances in Bioengineering and Clinical Engineering by Springer Nature Switzerland in 2024, page: 395-40310.1007/978-3-031-61973-1_37
- 2.Author(s): Michael Dwyer, Niels Bergsland, Robert ZivadinovPublished in Modern Inference Based on Health-Related Markers by Elsevier in 2024, page: 377-39710.1016/b978-0-12-815247-8.00014-0
- 3.Author(s): Devesh Singh, Martin DyrbaPublished in Informatik aktuell, Bildverarbeitung für die Medizin 2024 by Springer Fachmedien Wiesbaden in 2024, page: 149-15410.1007/978-3-658-44037-4_43
- 4.Author(s): Anne-Marie Rickmann, Fabian Bongratz, Christian WachingerPublished in Lecture Notes in Computer Science, Medical Image Computing and Computer Assisted Intervention – MICCAI 2023 by Springer Nature Switzerland in 2023, page: 318-32710.1007/978-3-031-43993-3_31
- 5.Author(s): Zhengwang Wu, Jiale Cheng, Fenqiang Zhao, Ya Wang, Yue Sun, Dajiang Zhu, Tianming Liu, Valerie Jewells, Weili Lin, Li Wang, Gang LiPublished in Lecture Notes in Computer Science, Medical Image Computing and Computer Assisted Intervention – MICCAI 2023 by Springer Nature Switzerland in 2023, page: 429-43810.1007/978-3-031-43993-3_42
- 6.Author(s): Richard McKinley, Christian RummelPublished in Lecture Notes in Computer Science, Medical Image Computing and Computer Assisted Intervention – MICCAI 2023 by Springer Nature Switzerland in 2023, page: 730-73910.1007/978-3-031-43999-5_69
- 7.Author(s): Antoine Legouhy, Ross Callaghan, Hojjat Azadbakht, Hui ZhangPublished in Lecture Notes in Computer Science, Information Processing in Medical Imaging by Springer Nature Switzerland in 2023, page: 614-62510.1007/978-3-031-34048-2_47
- 8.Author(s): Marc Modat, David M. Cash, Liane Dos Santos Canas, Martina Bocchetta, Sébastien OurselinPublished in Neuromethods, Machine Learning for Brain Disorders by Springer US in 2023, page: 807-84610.1007/978-1-0716-3195-9_25
- 9.Author(s): Xin Wang, Zhenrong Shen, Zhiyun Song, Sheng Wang, Mengjun Liu, Lichi Zhang, Kai Xuan, Qian WangPublished in Lecture Notes in Computer Science, Machine Learning in Medical Imaging by Springer Nature Switzerland in 2023, page: 23-3210.1007/978-3-031-45673-2_3
- 10.Author(s): Sadhana Ravikumar, Ranjit Ittyerah, Sydney Lim, Long Xie, Sandhitsu Das, Pulkit Khandelwal, Laura E. M. Wisse, Madigan L. Bedard, John L. Robinson, Terry Schuck, Murray Grossman, John Q. Trojanowski, Edward B. Lee, M. Dylan Tisdall, Karthik Prabhakaran, John A. Detre, David J. Irwin, Winifred Trotman, Gabor Mizsei, Emilio Artacho-Pérula, Maria Mercedes Iñiguez de Onzono Martin, Maria del Mar Arroyo Jiménez, Monica Muñoz, Francisco Javier Molina Romero, Maria del Pilar Marcos Rabal, Sandra Cebada-Sánchez, José Carlos Delgado González, Carlos de la Rosa-Prieto, Marta Córcoles Parada, David A. Wolk, Ricardo Insausti, Paul A. YushkevichPublished in Lecture Notes in Computer Science, Information Processing in Medical Imaging by Springer Nature Switzerland in 2023, page: 692-70410.1007/978-3-031-34048-2_53
- 11.Author(s): Stefanos Ioannou, Hana Chockler, Alexander Hammers, Andrew P. King, for the Alzheimer’s Disease Neuroimaging InitiativePublished in Lecture Notes in Computer Science, Machine Learning in Clinical Neuroimaging by Springer Nature Switzerland in 2022, page: 13-2210.1007/978-3-031-17899-3_2
- 12.Author(s): Fenqiang Zhao, Zhengwang Wu, Li Wang, Weili Lin, Gang LiPublished in Lecture Notes in Computer Science, Medical Image Computing and Computer Assisted Intervention – MICCAI 2022 by Springer Nature Switzerland in 2022, page: 163-17310.1007/978-3-031-16446-0_16
- 13.Author(s): Rodrigo Santa Cruz, Léo Lebrat, Darren Fu, Pierrick Bourgeat, Jurgen Fripp, Clinton Fookes, Olivier SalvadoPublished in Lecture Notes in Computer Science, Medical Image Computing and Computer Assisted Intervention – MICCAI 2022 by Springer Nature Switzerland in 2022, page: 496-50510.1007/978-3-031-16443-9_48
- 14.Author(s): Yuyan Ge, Zhenyu Tang, Lei Ma, Caiwen Jiang, Feng Shi, Shaoyi Du, Dinggang ShenPublished in Lecture Notes in Computer Science, Machine Learning in Medical Imaging by Springer Nature Switzerland in 2022, page: 466-47510.1007/978-3-031-21014-3_48
- 15.Author(s): Leonie Henschel, David Kügler, Derek S. Andrews, Christine W. Nordahl, Martin ReuterPublished in Lecture Notes in Computer Science, Medical Image Computing and Computer Assisted Intervention – MICCAI 2022 by Springer Nature Switzerland in 2022, page: 350-35910.1007/978-3-031-16443-9_34
- 16.Author(s): Anne-Marie Rickmann, Fabian Bongratz, Sebastian Pölsterl, Ignacio Sarasua, Christian WachingerPublished in Lecture Notes in Computer Science, Machine Learning in Clinical Neuroimaging by Springer Nature Switzerland in 2022, page: 3-1210.1007/978-3-031-17899-3_1
- 17.Author(s): Karthik Gopinath, Christian Desrosiers, Herve LombaertPublished in Lecture Notes in Computer Science, Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 by Springer International Publishing in 2021, page: 650-65910.1007/978-3-030-87234-2_61
- 18.Author(s): Christian Ewert, David Kügler, Anastasia Yendiki, Martin ReuterPublished in Mathematics and Visualization, Computational Diffusion MRI by Springer International Publishing in 2021, page: 81-9310.1007/978-3-030-73018-5_7
- 19.Author(s): Filip Rusak, Rodrigo Santa Cruz, Elliot Smith, Jurgen Fripp, Clinton Fookes, Pierrick Bourgeat, Andrew BradleyPublished in Lecture Notes in Computer Science, Simulation and Synthesis in Medical Imaging by Springer International Publishing in 2021, page: 3-1310.1007/978-3-030-87592-3_1
- 20.Author(s): Qiang Ma, Emma C. Robinson, Bernhard Kainz, Daniel Rueckert, Amir AlansaryPublished in Lecture Notes in Computer Science, Machine Learning in Clinical Neuroimaging by Springer International Publishing in 2021, page: 73-8110.1007/978-3-030-87586-2_8
- 21.Author(s): Leonie Henschel, Sailesh Conjeti, Santiago Estrada, Kersten Diers, Bruce Fischl, Martin ReuterPublished in Informatik aktuell, Bildverarbeitung für die Medizin 2020 by Springer Fachmedien Wiesbaden in 2020, page: 208-20810.1007/978-3-658-29267-6_46
- 1.Author(s): Pablo Laso, Stefano Cerri, Annabel Sorby-Adams, Jennifer Guo, Farrah Mateen, Philipp Goebl, Jiaming Wu, Peirong Liu, Hongwei Bran Li, Sean I. Young, Benjamin Billot, Oula Puonti, Gordon Sze, Sam Payabavash, Adam DeHavenon, Kevin N. Sheth, Matthew S. Rosen, John Kirsch, Nicola Strisciuglio, Jelmer M. Wolterink, Arman Eshaghi, Frederik Barkhof, W. Taylor Kimberly, Juan Eugenio IglesiasPublished in 2024 IEEE International Symposium on Biomedical Imaging (ISBI) by IEEE in 2024, page: 1-510.1109/isbi56570.2024.10635502
- 2.Author(s): Mohamed Khudri, Mostafa Abdelrahim, Mohamed T. Ali, Yaser A. Elnakib, Ahmed Shalaby, Ali Mahmoud, Sohail Contractor, Gregory N. Barnes, Ayman El-BazPublished in 2024 IEEE International Symposium on Biomedical Imaging (ISBI) by IEEE in 2024, page: 1-410.1109/isbi56570.2024.10635720
- 3.Author(s): Solveig K. Hammonds, Trygve Eftestøl, Ketil Oppedal, Alvaro Fernandez-QuilezPublished in 2024 4th International Conference on Applied Artificial Intelligence (ICAPAI) by IEEE in 2024, page: 1-710.1109/icapai61893.2024.10541140
- 4.Author(s): Mohamed Masoud, Pratyush Reddy, Farfalla Hu, Sergey PlisPublished in 2024 IEEE International Symposium on Biomedical Imaging (ISBI) by IEEE in 2024, page: 1-510.1109/isbi56570.2024.10635670
- 5.Author(s): Shruti P. Gadewar, Alyssa H. Zhu, Iyad Ba Gari, Sunanda Somu, Sophia I. Thomopoulos, Paul M. Thompson, Talia M. Nir, Neda JahanshadPublished in 2024 IEEE International Symposium on Biomedical Imaging (ISBI) by IEEE in 2024, page: 1-510.1109/isbi56570.2024.10635691
- 6.Author(s): Shruti P. Gadewar, Elnaz Nourollahimoghadam, Ravi R. Bhatt, Abhinaav Ramesh, Shayan Javid, Iyad Ba Gari, Alyssa H. Zhu, Sophia Thomopoulos, Paul M. Thompson, Neda JahanshadPublished in 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) by IEEE in 2023, page: 1-410.1109/embc40787.2023.10340442
- 7.Author(s): Camilo Laiton-Bonadiez, Germán Sanchez-Torres, Nallig Leal, Aura Polo-LlanosPublished in Memorias de la Conferencia Iberoamericana en Sistemas, Cibernética e Informática, Memorias de la Vigésima Segunda Conferencia Iberoamericana en Sistemas, Cibernética e Informática: CISCI 2023 by International Institute of Informatics and Cybernetics in 2023, page: 16-2010.54808/cisci2023.01.16
- 8.Author(s): Kai Tzu-Iunn Ong, Hana Kim, Minjin Kim, Jinseong Jang, Beomseok Sohn, Yoon Seong Choi, Dosik Hwang, Seong Jae Hwang, Jinyoung YeoPublished in 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI) by IEEE in 2023, page: 1-510.1109/isbi53787.2023.10230842
- 9.Author(s): Zexuan Wang, Jiong Chen, Wenxi Yang, Sumita Gara, Frederick Xu, Junhao Wen, Christos Davatzikos, Li ShenPublished in Medical Imaging 2023: Image Processing by SPIE in 2023, page: 12410.1117/12.2654399
- 10.Author(s): Yanwu Yang, Xutao Guo, Zhikai Chang, Chenfei Ye, Yang Xiang, Haiyan Lv, Ting MaPublished in 2022 IEEE International Conference on Image Processing (ICIP) by IEEE in 2022, page: 56-6010.1109/icip46576.2022.9897454
- 11.Author(s): Jayesh Kumar, Chinmay Satish Raut, Niravkumar PatelPublished in 2022 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS) by IEEE in 2022, page: 4018-402310.1109/iros47612.2022.9981164
- 12.Author(s): Sara Atek, Imane Mehidi, Dalel Jabri, Djamel E. C. BelkhiatPublished in 2022 7th International Conference on Image and Signal Processing and their Applications (ISPA) by IEEE in 2022, page: 1-610.1109/ispa54004.2022.9786367
- 13.Author(s): Raissa Souza, Agampreet Aulakh, Pauline Mouches, Anup Tuladhar, Matthias Wilms, Sönke Langner, Nils D. ForkertPublished in Medical Imaging 2022: Imaging Informatics for Healthcare, Research, and Applications by SPIE in 2022, page: 110.1117/12.2612728
- 14.Author(s): Mythri V, Alphin J Thottupattu, Naren Akash R J, Jayanthi SivaswamyPublished in 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI) by IEEE in 2022, page: 1-410.1109/isbi52829.2022.9761673
- 15.Author(s): Khosro Rezaee, Mohammad R. Khosravi, Mohammad Sabri, Khaled Al-QawasmiPublished in 2022 International Engineering Conference on Electrical, Energy, and Artificial Intelligence (EICEEAI) by IEEE in 202210.1109/eiceeai56378.2022.10050454
- 16.Author(s): Ashkan Nejad, Saeed Masoudnia, Mohammad-Reza Nazem-ZadehPublished in 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) by IEEE in 2022, page: 2140-214310.1109/embc48229.2022.9871715
- 17.Author(s): Marius Schmidt-Mengin, Vito A. G. Ricigliano, Benedetta Bodini, Emanuele Morena, Annalisa Colombi, Maryem Hamzaoui, Arya Yazdan Panah, Bruno Stankoff, Olivier ColliotPublished in Medical Imaging 2022: Image Processing by SPIE in 2022, page: 610.1117/12.2612912
- 18.Author(s): Fabian Bongratz, Anne-Marie Rickmann, Sebastian Polsterl, Christian WachingerPublished in 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) by IEEE in 2022, page: 20741-2075110.1109/cvpr52688.2022.02011
- 19.Author(s): Mujun An, Congcong Wang, Kemeng Tao, Jing Han, Kun Zhao, Zhi LiuPublished in 2022 15th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI) by IEEE in 2022, page: 1-610.1109/cisp-bmei56279.2022.9980286
- 20.Author(s): Shruti Gadewar, Alyssa H. Zhu, Sophia I. Thomopoulos, Zhuocheng Li, Iyad Ba Gari, Piyush Maiti, Paul M. Thompson, Neda JahanshadPublished in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) by IEEE in 2021, page: 1288-129110.1109/isbi48211.2021.9433755
- 21.Author(s): Salamata Konate, Leo Lebrat, Rodrigo Santa Cruz, Pierrick Bourgeat, Vincent Dore, Jurgen Fripp, Andrew Bradley, Clinton Fookes, Olivier SalvadoPublished in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) by IEEE in 2021, page: 362-36610.1109/isbi48211.2021.9433972
- 22.Author(s): Rodrigo Santa Cruz, Leo Lebrat, Pierrick Bourgeat, Vincent Dore, Jason Dowling, Jurgen Fripp, Clinton Fookes, Olivier SalvadoPublished in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) by IEEE in 2021, page: 711-71510.1109/isbi48211.2021.9434039
- 23.Author(s): Anqi Yang, Feng Pan, Vishwanath Saragadam, Duy Dao, Zhuo Hui, Jen-Hao Rick Chang, Aswin C. SankaranarayananPublished in 2021 IEEE Winter Conference on Applications of Computer Vision (WACV) by IEEE in 2021, page: 335-34410.1109/wacv48630.2021.00038
- 24.Author(s): Rodrigo Santa Cruz, Leo Lebrat, Pierrick Bourgeat, Clinton Fookes, Jurgen Fripp, Olivier SalvadoPublished in 2021 IEEE Winter Conference on Applications of Computer Vision (WACV) by IEEE in 202110.1109/wacv48630.2021.00085
- 25.Author(s): Louise Bloch, Christoph M. FriedrichPublished in 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS) by IEEE in 2021, page: 113-11810.1109/cbms52027.2021.00096
- 26.Author(s): Leonie Henschel, Sailesh Conjeti, Santiago Estrada, Kersten Diers, Bruce Fischl, Martin ReuterPublished by Organization of Human Brain Mapping in 2020, page: 100
- 1.Author(s): Alex Fedorov, Eloy Geenjaar, Lei Wu, Tristan Sylvain, Thomas P. DeRamus, Margaux Luck, Maria Misiura, Girish Mittapalle, R. Devon Hjelm, Sergey M. Plis, Vince D. CalhounPublished in NeuroImage by Elsevier BV in 2024, page: 12048510.1016/j.neuroimage.2023.120485
- 2.Author(s): Poonam Rani Verma, Ashish Kumar BhandariPublished in Biomedical Signal Processing and Control by Elsevier BV in 2024, page: 10565810.1016/j.bspc.2023.105658
- 3.Author(s): Sylvain Gerbaud, Arthur Cavalier, Sébastien Horna, Rita Zrour, Mathieu Naudin, Carole Guillevin, Philippe MeseurePublished in Computers & Graphics by Elsevier BV in 2024, page: 10394710.1016/j.cag.2024.103947
- 4.Author(s): Michele Svanera, Mattia Savardi, Alberto Signoroni, Sergio Benini, Lars MuckliPublished in Medical Image Analysis by Elsevier BV in 2024, page: 10309010.1016/j.media.2024.103090
- 5.Author(s): Chaokai Zhang, Lara Bartels, Adam Clansey, Julian Kloiber, Daniel Bondi, Paul van Donkelaar, Lyndia Wu, Alexander Rauscher, Songbai JiPublished in Computers in Biology and Medicine by Elsevier BV in 2024, page: 10810910.1016/j.compbiomed.2024.108109
- 6.Author(s): Limei Wang, Yue Sun, Weili Lin, Gang Li, Li WangPublished in Intelligent Medicine by Elsevier BV in 2024, page: 65-7410.1016/j.imed.2023.05.002
- 7.Author(s): Lukas Fisch, Stefan Zumdick, Carlotta Barkhau, Daniel Emden, Jan Ernsting, Ramona Leenings, Kelvin Sarink, Nils R. Winter, Benjamin Risse, Udo Dannlowski, Tim HahnPublished in Computers in Biology and Medicine by Elsevier BV in 2024, page: 10884510.1016/j.compbiomed.2024.108845
- 8.Author(s): Heath M. Lorzel, Mark D. AllenPublished in NeuroImage by Elsevier BV in 2024, page: 12050510.1016/j.neuroimage.2023.120505
- 9.Author(s): Fabian Bongratz, Anne-Marie Rickmann, Christian WachingerPublished in Medical Image Analysis by Elsevier BV in 2024, page: 10309310.1016/j.media.2024.103093
- 10.Author(s): Jose Gomez-Tames, Mariano Fernández-CorazzaPublished in Journal of Clinical Medicine by MDPI AG in 2024, page: 308410.3390/jcm13113084
- 11.Author(s): Thiago J.R. Rezende, Emilien Petit, Young Woo Park, Sophie Tezenas du Montcel, James M. Joers, Jonathan M. DuBois, H. Moore Arnold, Michal Povazan, Guita Banan, Romain Valabregue, Philipp Ehses, Jennifer Faber, Pierrick Coupé, Chiadi U. Onyike, Peter B. Barker, Jeremy D. Schmahmann, Eva‐Maria Ratai, Sub H. Subramony, Thomas H. Mareci, Khalaf O. Bushara, Henry Paulson, Thomas Klockgether, Alexandra Durr, Tetsuo Ashizawa, Christophe Lenglet, Gülin Öz, READISCA ConsortiumPublished in Movement Disorders by Wiley in 2024, page: 1856-186710.1002/mds.29934
- 12.Author(s): Laurien De Roeck, Jeroen Blommaert, Patrick Dupont, Stefan Sunaert, Charlotte Sleurs, Maarten LambrechtPublished in Scientific Reports by Springer Science and Business Media LLC in 202410.1038/s41598-024-63716-2
- 13.Author(s): Hiromasa Takemura, John A. Kruper, Toshikazu Miyata, Ariel RokemPublished in Magnetic Resonance in Medical Sciences by Japanese Society for Magnetic Resonance in Medicine in 2024, page: 316-34010.2463/mrms.rev.2024-0007
- 14.Author(s): Hao Niu, Yuxiang Zhou, Xiaohao Yan, Jun Wu, Yuncheng Shen, Zhang Yi, Junjie HuPublished in Artificial Intelligence Review by Springer Science and Business Media LLC in 202410.1007/s10462-024-10894-0
- 15.Author(s): Haoteng Tang , Siyuan Dai, Eric M. Zou, Guodong Liu, Ryan Ahearn, Ryan Krafty, Michel Modo, Liang ZhanPublished in Mathematics by MDPI AG in 2024, page: 94010.3390/math12070940
- 16.Author(s): Monique Tourell, Jin Jin, Beata Bachrata, Ashley Stewart, Stefan Ropele, Christian Enzinger, Saskia Bollmann, Steffen Bollmann, Simon Daniel Robinson, Kieran O'Brien, Markus BarthPublished in Magnetic Resonance in Medicine by Wiley in 2024, page: 997-101010.1002/mrm.30101
- 17.Author(s): Yeong-Jae Jeon, Shin-Eui Park, Hyeon-Man BaekPublished in Brain Sciences by MDPI AG in 2024, page: 40110.3390/brainsci14040401
- 18.Author(s): Cyrus A. Raji, Somayeh Meysami, Sam Hashemi, Saurabh Garg, Nasrin Akbari, Gouda Ahmed, Yosef Gavriel Chodakiewitz, Thanh Duc Nguyen, Kellyann Niotis, David A. Merrill, Rajpaul AttariwalaPublished in Journal of Alzheimer's Disease by SAGE Publications in 2024, page: 829-83910.3233/jad-230740
- 19.Author(s): Carolin Gaiser, Rick van der Vliet, Augustijn A. A. de Boer, Opher Donchin, Pierre Berthet, Gabriel A. Devenyi, M. Mallar Chakravarty, Jörn Diedrichsen, Andre F. Marquand, Maarten A. Frens, Ryan L. MuetzelPublished in Nature Communications by Springer Science and Business Media LLC in 202410.1038/s41467-024-46398-2
- 20.Author(s): Hampus Olsson, Jason Michael Millward, Ludger Starke, Thomas Gladytz, Tobias Klein, Jana Fehr, Wei-Chang Lai, Christoph Lippert, Thoralf Niendorf, Sonia WaicziesPublished in PLOS ONE by Public Library of Science (PLoS) in 2024, page: e030113210.1371/journal.pone.0301132
- 21.Author(s): Albert Clèrigues, Sergi Valverde, Arnau Oliver, Xavier LladóPublished in Computers in Biology and Medicine by Elsevier BV in 2024, page: 10881110.1016/j.compbiomed.2024.108811
- 22.Author(s): Difei Wang, Rüdiger Stirnberg, Tony StöckerPublished in Magnetic Resonance in Medicine by Wiley in 202410.1002/mrm.30217
- 23.Author(s): Peter C Van Dyken, Michael MacKinley, Ali R Khan, Lena PalaniyappanPublished in Schizophrenia Bulletin Open by Oxford University Press (OUP) in 202410.1093/schizbullopen/sgae010
- 24.Author(s): Andre A. Payman, Ivan El-Sayed, Roberto Rodriguez RubioPublished in World Neurosurgery by Elsevier BV in 2024, page: e403-e41010.1016/j.wneu.2024.08.137
- 25.Author(s): Jiaxin Li, Yueqin Hu, Yunzhi Xu, Xue Feng, Craig H. Meyer, Weiying Dai, Li Zhao, for the Alzheimer’s Disease Neuroimaging InitiativePublished in Fluids and Barriers of the CNS by Springer Science and Business Media LLC in 202410.1186/s12987-024-00554-4
- 26.Author(s): Xin Wang, Sheng Wang, Honglin Xiong, Kai Xuan, Zixu Zhuang, Mengjun Liu, Zhenrong Shen, Xiangyu Zhao, Lichi Zhang, Qian WangPublished in Medical Image Analysis by Elsevier BV in 2024, page: 10315810.1016/j.media.2024.103158
- 27.Author(s): Arnaud Attyé, Félix Renard, Vanina Anglade, Alexandre Krainik, Philippe Kahane, Boris Mansencal, Pierrick Coupé, Fernando CalamantePublished in Scientific Reports by Springer Science and Business Media LLC in 202410.1038/s41598-024-58141-4
- 28.Author(s): Yeong Sim Choe, Regina E.Y. Kim, Hye Weon Kim, JeeYoung Kim, Hyunji Lee, Min Kyoung Lee, Minho Lee, Keun You Kim, Se-Hong Kim, Ji-hoon Kim, Jun-Young Lee, Eosu Kim, Donghyeon Kim, Hyun Kook LimPublished in Journal of Alzheimer's Disease Reports by SAGE Publications in 2024, page: 863-87610.3233/adr-230105
- 29.Author(s): Alfredo Lucas, Andrew Revell, Kathryn A. DavisPublished in Nature Reviews Neurology by Springer Science and Business Media LLC in 2024, page: 319-33610.1038/s41582-024-00965-9
- 30.Author(s): Manjunath Ramanna Lamani, Julian Benadit PernabasPublished in Recent Advances in Computer Science and Communications by Bentham Science Publishers Ltd. in 202410.2174/0126662558284886240130154414
- 31.Author(s): Lars Smolders, Wouter De Baene, Geert-Jan Rutten, Remco van der Hofstad, Luc FlorackPublished in Brain Structure and Function by Springer Science and Business Media LLC in 2024, page: 1209-122310.1007/s00429-024-02796-2
- 32.Author(s): Lachlan T. Strike, Rebecca Kerestes, Katie L. McMahon, Greig I. de Zubicaray, Ian H. Harding, Sarah E. MedlandPublished in Human Brain Mapping by Wiley in 202410.1002/hbm.26717
- 33.Author(s): Kirill Sobyanin, Sofya KulikovaPublished in Informatics and Automation, Информатика и автоматизация by SPIIRAS in 2024, page: 1022-104610.15622/ia.23.4.4
- 34.Author(s): Jaime Simarro, Maria Ines Meyer, Simon Van Eyndhoven, Thanh Vân Phan, Thibo Billiet, Diana M. Sima, Els OrtibusPublished in Scientific Reports by Springer Science and Business Media LLC in 202410.1038/s41598-024-61798-6
- 35.Author(s): Fan Zhang, Kang Ik Kevin Cho, Johanna Seitz-Holland, Lipeng Ning, Jon Haitz Legarreta, Yogesh Rathi, Carl-Fredrik Westin, Lauren J. O’Donnell, Ofer PasternakPublished in IEEE Transactions on Medical Imaging by Institute of Electrical and Electronics Engineers (IEEE) in 2024, page: 1191-120210.1109/tmi.2023.3331691
- 36.Author(s): Ann-Marie G. de Lange, Esten H. Leonardsen, Claudia Barth, Louise S. Schindler, Arielle Crestol, Madelene C. Holm, Sivaniya Subramaniapillai, Dónal Hill, Dag Alnæs, Lars T. WestlyePublished in Psychoneuroendocrinology by Elsevier BV in 2024, page: 10704010.1016/j.psyneuen.2024.107040
- 37.Author(s): Chundan Xu, Jie Li, Yakui Wang, Lixue Wang, Yizhe Wang, Xiaofeng Zhang, Weiqi Liu, Jingang Chen, Aleksandra Vatian, Natalia Gusarova, Chuyang Ye, Zhuozhao ZhengPublished in NeuroImage by Elsevier BV in 2024, page: 12081210.1016/j.neuroimage.2024.120812
- 38.Author(s): Felix Bitzer, Lennart Walger, Tobias Bauer, Freya Schulte, Florian C. Gaertner, Matthias Schmitz, Martin Schidlowski, Randi von Wrede, Attila Rácz, Tobias Baumgartner, Vadym Gnatkovsky, Daniel Paech, Valeri Borger, Hartmut Vatter, Bernd Weber, Dominik L. Michels, Tony Stöcker, Markus Essler, Josemir W. Sander, Alexander Radbruch, Rainer Surges, Theodor RüberPublished in Annals of Neurology by Wiley in 2024, page: 1160-117310.1002/ana.27061
- 39.Author(s): Eya Khadhraoui, Thomas Nickl-Jockschat, Hans Henkes, Daniel Behme, Sebastian Johannes MüllerPublished in Frontiers in Aging Neuroscience by Frontiers Media SA in 202410.3389/fnagi.2024.1459652
- 40.Author(s): Hunju Lee, Sang Yeol Yong, Hyowon Choi, Ga Young Yoon, Sangbaek KohPublished in Frontiers in Aging Neuroscience by Frontiers Media SA in 202410.3389/fnagi.2024.1389476
- 41.Author(s): Louise Bloch, Christoph M. FriedrichPublished in Computers in Biology and Medicine by Elsevier BV in 2024, page: 10802910.1016/j.compbiomed.2024.108029
- 42.Author(s): Iman Aganj, Jocelyn Mora, Bruce Fischl, Jean C. AugustinackPublished in Frontiers in Neuroscience by Frontiers Media SA in 202410.3389/fnins.2024.1375530
- 43.Author(s): Valentina Visani, Mattia Veronese, Francesca B. Pizzini, Annalisa Colombi, Valerio Natale, Corina Marjin, Agnese Tamanti, Julia J. Schubert, Noha Althubaity, Inés Bedmar-Gómez, Neil A. Harrison, Edward T. Bullmore, Federico E. Turkheimer, Massimiliano Calabrese, Marco CastellaroPublished in Computers in Biology and Medicine by Elsevier BV in 2024, page: 10916410.1016/j.compbiomed.2024.109164
- 44.Author(s): Ahmed Mohamed Radwan, Louise Emsell, Kristof Vansteelandt, Evy Cleeren, Ronald Peeters, Steven De Vleeschouwer, Tom Theys, Patrick Dupont, Stefan SunaertPublished in Human Brain Mapping by Wiley in 202410.1002/hbm.26662
- 45.Author(s): Jong Sung Park, Shreyas Fadnavis, Eleftherios GaryfallidisPublished in Communications Medicine by Springer Science and Business Media LLC in 202410.1038/s43856-024-00452-8
- 46.Author(s): David Romascano, Michael Rebsamen, Piotr Radojewski, Timo Blattner, Richard McKinley, Roland Wiest, Christian RummelPublished in NeuroImage: Clinical by Elsevier BV in 2024, page: 10362410.1016/j.nicl.2024.103624
- 47.Author(s): Dmitrii Lachinov, Arunava Chakravarty, Christoph Grechenig, Ursula Schmidt-Erfurth, Hrvoje BogunovićPublished in IEEE Transactions on Medical Imaging by Institute of Electrical and Electronics Engineers (IEEE) in 2024, page: 1165-117910.1109/tmi.2023.3330576
- 48.Author(s): Sonia Ben Hassen, Mohamed Neji, Zain Hussain, Amir Hussain, Adel M. Alimi, Mondher FrikhaPublished in Neurocomputing by Elsevier BV in 2024, page: 12732510.1016/j.neucom.2024.127325
- 49.Author(s): Thanuja Dharmadasa, Nathan Pavey, Sicong Tu, Parvathi Menon, William Huynh, Colin J. Mahoney, Hannah C. Timmins, Mana Higashihara, Mehdi van den Bos, Kazumoto Shibuya, Satoshi Kuwabara, Julian Grosskreutz, Matthew C. Kiernan, Steve VucicPublished in Clinical Neurophysiology by Elsevier BV in 2024, page: 68-8910.1016/j.clinph.2024.04.010
- 50.Author(s): Junmo Kwon, Jonghun Kim, Hyunjin ParkPublished in Computer Methods and Programs in Biomedicine by Elsevier BV in 2024, page: 10833810.1016/j.cmpb.2024.108338
- 1.Author(s): Leonie Henschel, David Kügler, Lilla Zollei, Martin ReuterPublished by OHBM 2023 in 2023
- 2.Author(s): Santiago Estrada, Emad Bahrami, David Kügler, Dilshad Mousa, Peng Xu, Monique Breteler, N. Ahmad Aziz, Martin ReuterPublished by Organization of Human Brain Mapping 2022 in 2022, page: 64
- 3.Author(s): Leonie Henschel, David Kügler, Martin ReuterPublished by Organization of Human Brain Mapping 2022 in 2022, page: 66
- 4.Author(s): Leonie Henschel, Sailesh Conjeti, Santiago Estrada, Kersten Diers, Bruce Fischl, Martin ReuterPublished by Organization of Human Brain Mapping 2020 in 2020, page: 100
- 5.Author(s): Santiago Estrada, Ran Lu, Kersten Diers, Weiyi Zeng, Tony Stöcker, Monique Breteler, Martin Reuter
